Learning transcriptome dynamics for discovery of optimal genetic reporters of novel compounds
Published in In Review, Nature Communications, 2022
Recommended citation: A. Hasnain, S. Balakrishnan, ..., E. Yeung. (2022). "Learning transcriptome dynamics for discovery of optimal genetic reporters of novel compounds." bioRXiv preprint https://doi.org/10.1101/2022.05.27.493781.
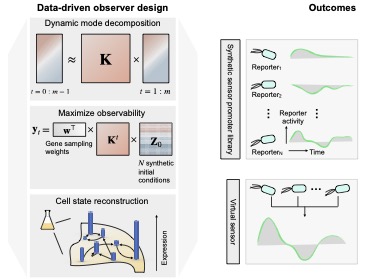
We develop a computational framework for the discovery of an optimal panel of reporters for novel compounds. The framework consists of learning a low-dimensional representation of transcriptome dynamics from time-course RNA-seq data. The low-dimensional representation is then used to identify a panel of genes that maximize a measure of observability of the dynamical system. A reporter library is constructed and characterized for the organophosphate malathion. We also explore the computational expressivity of the reporter library in an analog computing setting. Finally, we show the ability of the reporters to detect malathion in an environmental setting. Preprint on bioRXiv: